Visualising SARS-CoV-2 Spike Protein Mutations:
An Interactive Web-Based Application
My dissertation focused on visualising SARS-CoV-2 spike protein mutations using techniques in molecular visualisation, 3D modeling, animation, and interactive app development.
This project was conducted in association with The Glasgow School of Art, The University of Glasgow MRC-Centre for Virus Research, and the COVID-19 Genomics UK (COG-UK) Consortium.
This project received The Chairman’s Award from The Glasgow School of Art as the top project from the School of Simulation and Visualisation; read more here.
Press Releases:
3D Molecular Animations
Proteins were modeled using a variety of software. PyMOL and Chimera were used molecular visualisation and PDB generation, while molecular Maya and Autodesk Maya for 3D modeling and animation.
Here are a few quick sneak peeks into the animations:
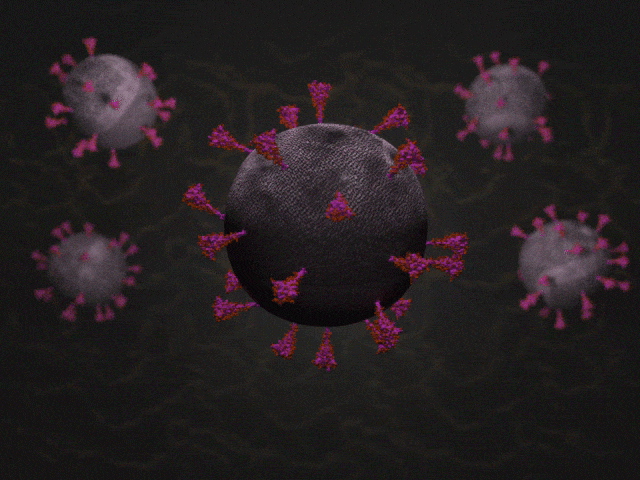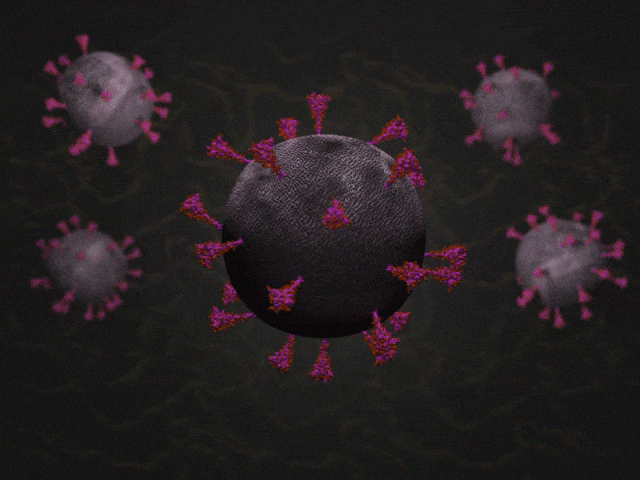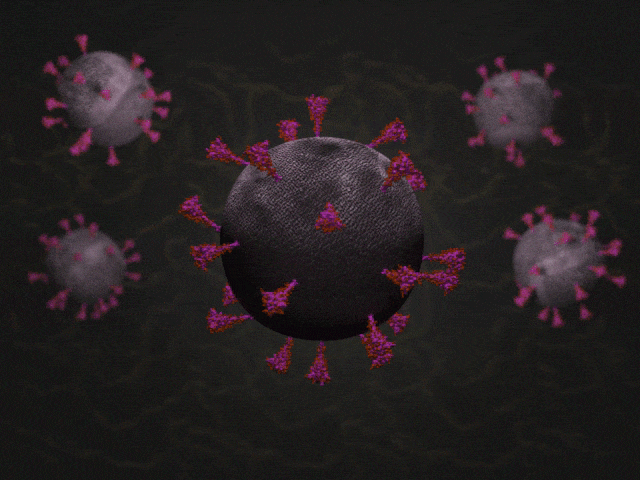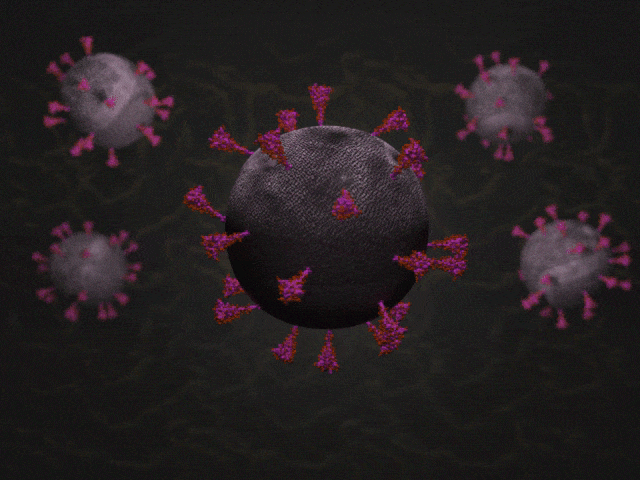
SARS-CoV-2 virions

Spike protein binding to ACE-2
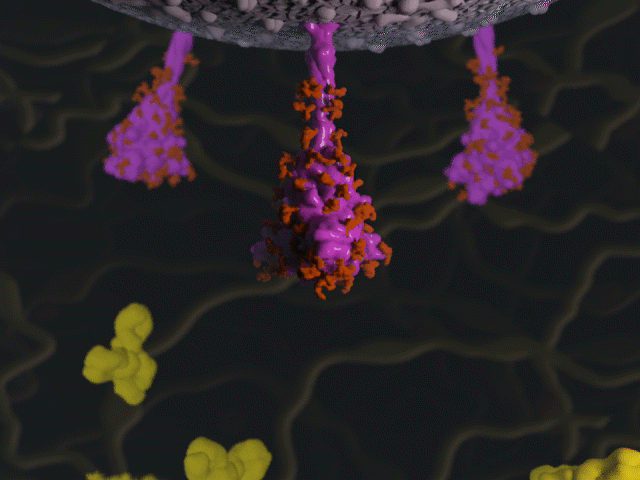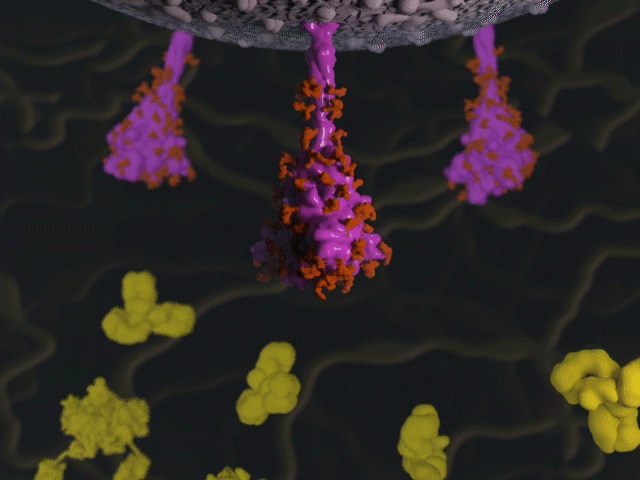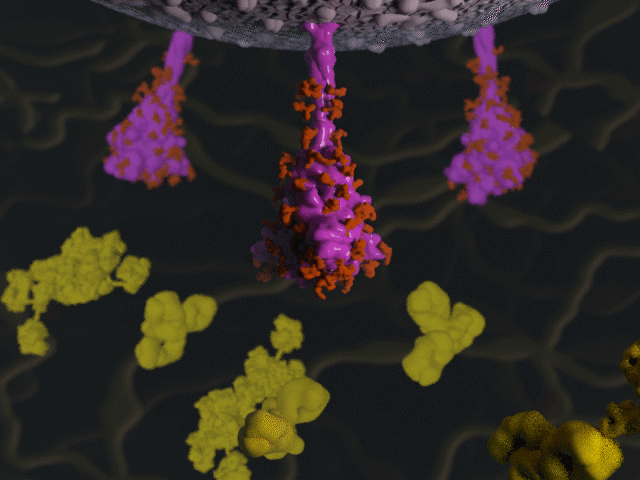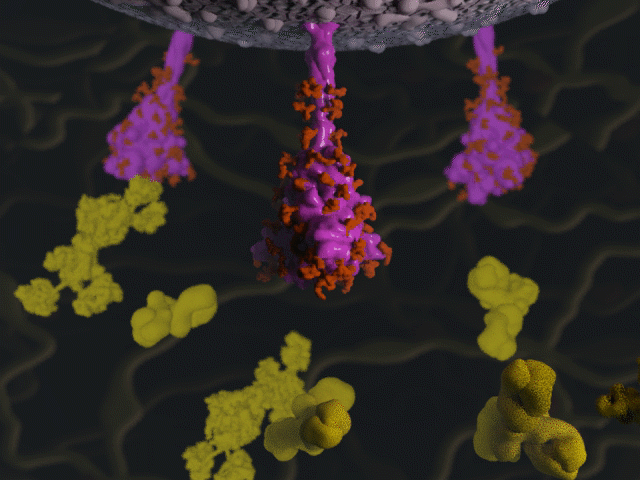
Antibodies & the spike protein
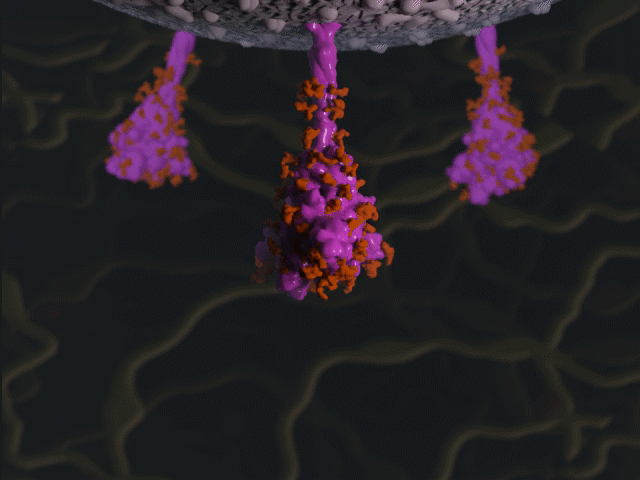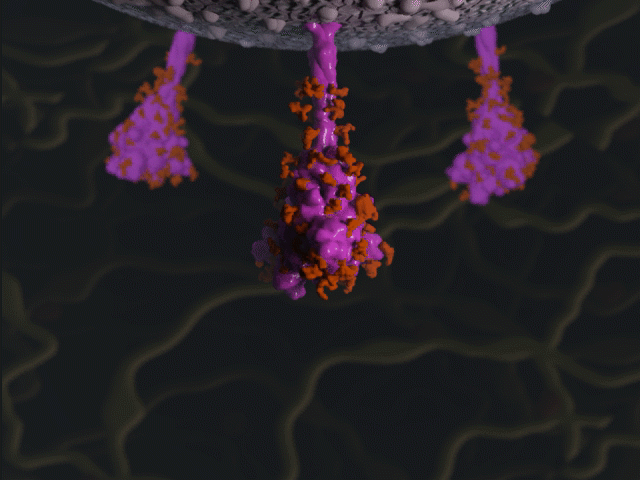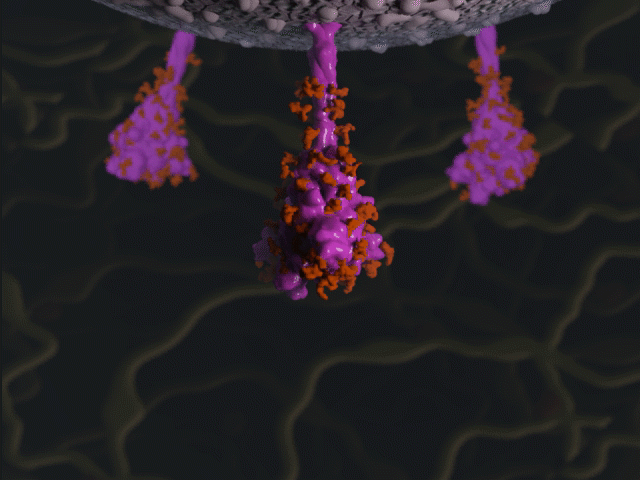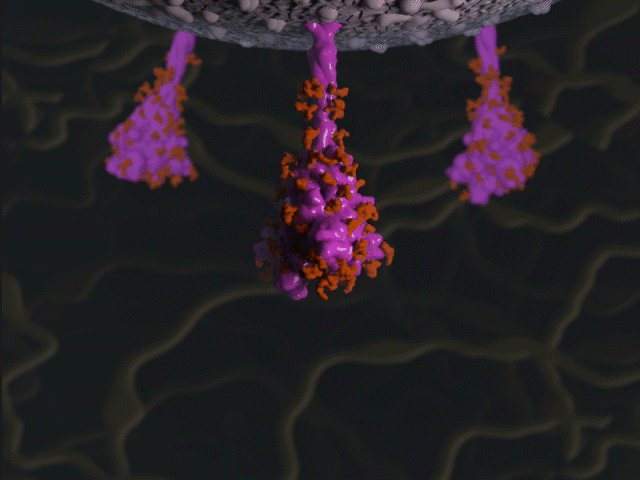
Spike protein glycosylation jiggle
Interactive App
The 3D models of the spike proteins were generated using the above software and imported into Unity, where the full application was developed from scratch.
You can trial the beta version of the app here: https://sc2-application.itch.io/sars-cov-2-mutation-explorer (please feel free to contact me with any feedback, positive or constructive!). The final updates and bug fixes will be implemented before official publishing.
Key features include:
An interactive 3D spike protein model with interactive regions of interest on the protein, including the Receptor-Binding Domain (RBD), N-Terminal Domain (NTD), Furin-Cleavage Site (FCS), and Glycosylation.
A Variants of Concern (VOC) menu, providing relevant up-to-date information on current VOCs, including the Delta and Omicron variants.
Region-specific information menus for the RBD, NTD, FCS, and Glycosylation; within the RBD and NTD menus are the 3D molecular animations shown above

Opening Scene

Instructions

Interactive 3D Spike Protein

Variants of Concern (VOC) Menu

RBD Information

NTD Information

FCS Information

Glycosylation Information